3. Linear Models for Regression¶
import numpy as np
from scipy.stats import multivariate_normal
import matplotlib.pyplot as plt
%matplotlib inline
from prml.preprocess import GaussianFeature, PolynomialFeature, SigmoidalFeature
from prml.linear import (
BayesianRegression,
EmpiricalBayesRegression,
LinearRegression,
RidgeRegression
)
np.random.seed(1234)
def create_toy_data(func, sample_size, std, domain=[0, 1]):
x = np.linspace(domain[0], domain[1], sample_size)
np.random.shuffle(x)
t = func(x) + np.random.normal(scale=std, size=x.shape)
return x, t
3.1 Linear Basis Function Models¶
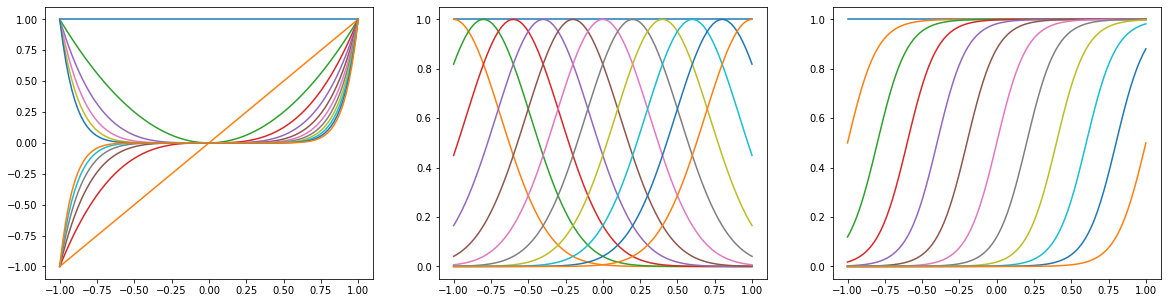
x = np.linspace(-1, 1, 100)
X_polynomial = PolynomialFeature(11).transform(x[:, None])
X_gaussian = GaussianFeature(np.linspace(-1, 1, 11), 0.1).transform(x)
X_sigmoidal = SigmoidalFeature(np.linspace(-1, 1, 11), 10).transform(x)
plt.figure(figsize=(20, 5))
for i, X in enumerate([X_polynomial, X_gaussian, X_sigmoidal]):
plt.subplot(1, 3, i + 1)
for j in range(12):
plt.plot(x, X[:, j])
3.1.1 Maximum likelihood and least squares¶
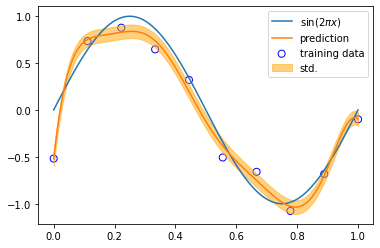
def sinusoidal(x):
return np.sin(2 * np.pi * x)
x_train, y_train = create_toy_data(sinusoidal, 10, 0.25)
x_test = np.linspace(0, 1, 100)
y_test = sinusoidal(x_test)
# Pick one of the three features below
# feature = PolynomialFeature(8)
feature = GaussianFeature(np.linspace(0, 1, 8), 0.1)
# feature = SigmoidalFeature(np.linspace(0, 1, 8), 10)
X_train = feature.transform(x_train)
X_test = feature.transform(x_test)
model = LinearRegression()
model.fit(X_train, y_train)
y, y_std = model.predict(X_test, return_std=True)
plt.scatter(x_train, y_train, facecolor="none", edgecolor="b", s=50, label="training data")
plt.plot(x_test, y_test, label="$\sin(2\pi x)$")
plt.plot(x_test, y, label="prediction")
plt.fill_between(
x_test, y - y_std, y + y_std,
color="orange", alpha=0.5, label="std.")
plt.legend()
plt.show()
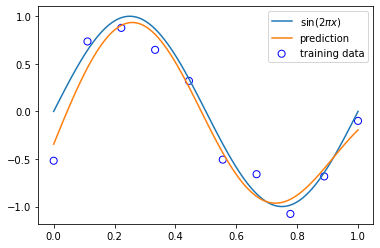
3.1.4 Regularized least squares¶
model = RidgeRegression(alpha=1e-3)
model.fit(X_train, y_train)
y = model.predict(X_test)
plt.scatter(x_train, y_train, facecolor="none", edgecolor="b", s=50, label="training data")
plt.plot(x_test, y_test, label="$\sin(2\pi x)$")
plt.plot(x_test, y, label="prediction")
plt.legend()
plt.show()
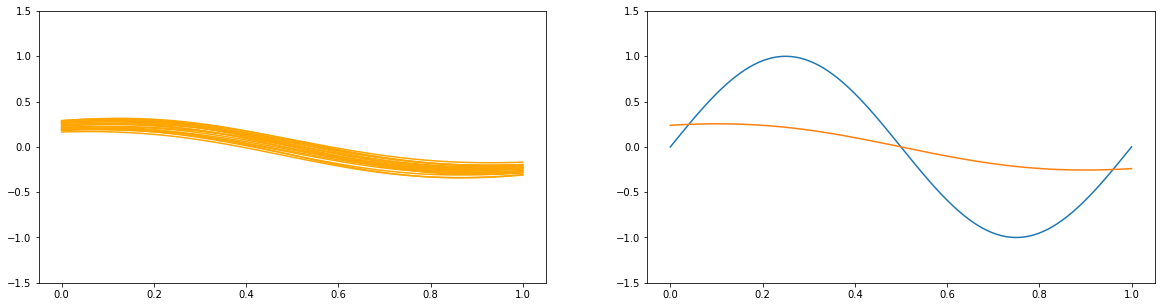
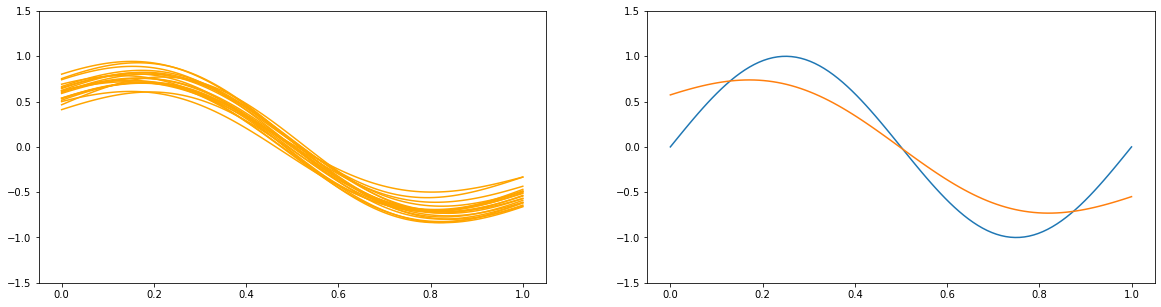
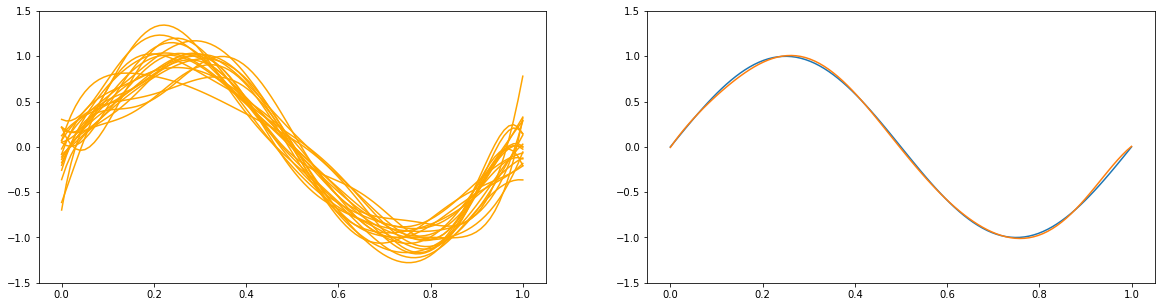
3.2 The Bias-Variance Decomposition¶
# feature = PolynomialFeature(24)
feature = GaussianFeature(np.linspace(0, 1, 24), 0.1)
# feature = SigmoidalFeature(np.linspace(0, 1, 24), 10)
for a in [1e2, 1., 1e-9]:
y_list = []
plt.figure(figsize=(20, 5))
plt.subplot(1, 2, 1)
for i in range(100):
x_train, y_train = create_toy_data(sinusoidal, 25, 0.25)
X_train = feature.transform(x_train)
X_test = feature.transform(x_test)
model = BayesianRegression(alpha=a, beta=1.)
model.fit(X_train, y_train)
y = model.predict(X_test)
y_list.append(y)
if i < 20:
plt.plot(x_test, y, c="orange")
plt.ylim(-1.5, 1.5)
plt.subplot(1, 2, 2)
plt.plot(x_test, y_test)
plt.plot(x_test, np.asarray(y_list).mean(axis=0))
plt.ylim(-1.5, 1.5)
plt.show()
3.3 Bayesian Linear Regression¶
3.3.1 Parameter distribution¶
def linear(x):
return -0.3 + 0.5 * x
x_train, y_train = create_toy_data(linear, 20, 0.1, [-1, 1])
x = np.linspace(-1, 1, 100)
w0, w1 = np.meshgrid(
np.linspace(-1, 1, 100),
np.linspace(-1, 1, 100))
w = np.array([w0, w1]).transpose(1, 2, 0)
feature = PolynomialFeature(degree=1)
X_train = feature.transform(x_train)
X = feature.transform(x)
model = BayesianRegression(alpha=1., beta=100.)
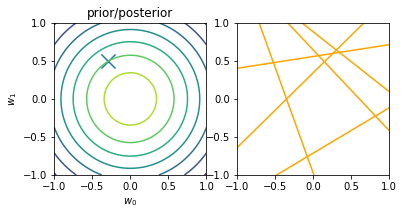
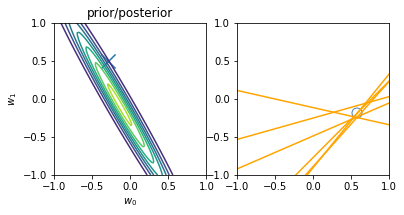
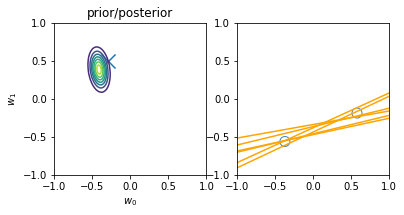
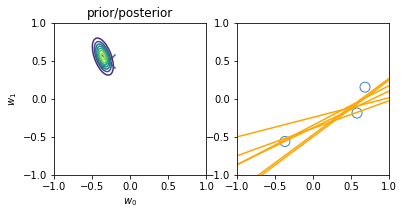
for begin, end in [[0, 0], [0, 1], [1, 2], [2, 3], [3, 20]]:
model.fit(X_train[begin: end], y_train[begin: end])
plt.subplot(1, 2, 1)
plt.scatter(-0.3, 0.5, s=200, marker="x")
plt.contour(w0, w1, multivariate_normal.pdf(w, mean=model.w_mean, cov=model.w_cov))
plt.gca().set_aspect('equal')
plt.xlabel("$w_0$")
plt.ylabel("$w_1$")
plt.title("prior/posterior")
plt.subplot(1, 2, 2)
plt.scatter(x_train[:end], y_train[:end], s=100, facecolor="none", edgecolor="steelblue", lw=1)
plt.plot(x, model.predict(X, sample_size=6), c="orange")
plt.xlim(-1, 1)
plt.ylim(-1, 1)
plt.gca().set_aspect('equal', adjustable='box')
plt.show()
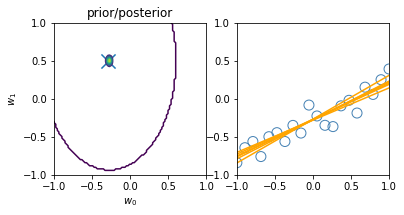
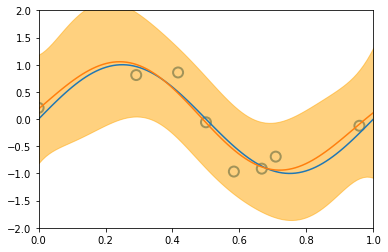
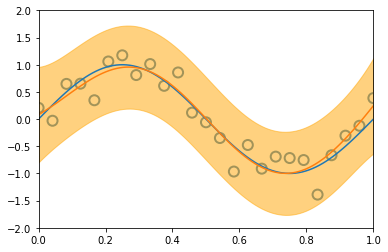
3.3.2 Predictive distribution¶
x_train, y_train = create_toy_data(sinusoidal, 25, 0.25)
x_test = np.linspace(0, 1, 100)
y_test = sinusoidal(x_test)
feature = GaussianFeature(np.linspace(0, 1, 9), 0.1)
X_train = feature.transform(x_train)
X_test = feature.transform(x_test)
model = BayesianRegression(alpha=1e-3, beta=2.)
for begin, end in [[0, 1], [1, 2], [2, 4], [4, 8], [8, 25]]:
model.fit(X_train[begin: end], y_train[begin: end])
y, y_std = model.predict(X_test, return_std=True)
plt.scatter(x_train[:end], y_train[:end], s=100, facecolor="none", edgecolor="steelblue", lw=2)
plt.plot(x_test, y_test)
plt.plot(x_test, y)
plt.fill_between(x_test, y - y_std, y + y_std, color="orange", alpha=0.5)
plt.xlim(0, 1)
plt.ylim(-2, 2)
plt.show()
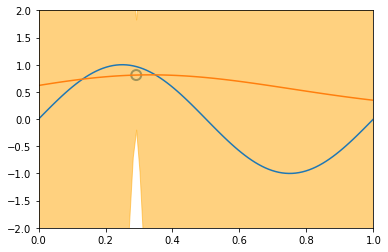
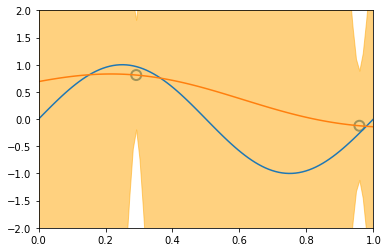
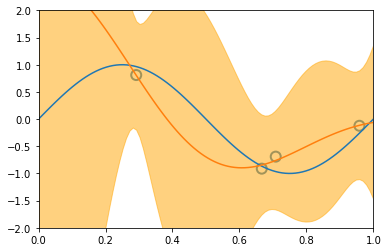
3.5 The Evidence Approximation¶
def cubic(x):
return x * (x - 5) * (x + 5)
x_train, y_train = create_toy_data(cubic, 30, 10, [-5, 5])
x_test = np.linspace(-5, 5, 100)
evidences = []
models = []
for i in range(8):
feature = PolynomialFeature(degree=i)
X_train = feature.transform(x_train)
model = EmpiricalBayesRegression(alpha=100., beta=100.)
model.fit(X_train, y_train, max_iter=100)
evidences.append(model.log_evidence(X_train, y_train))
models.append(model)
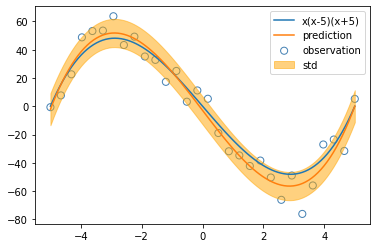
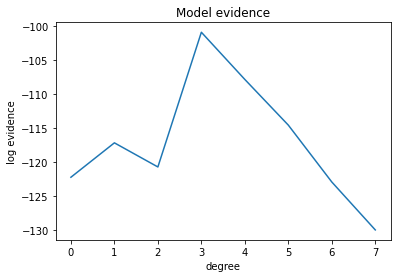
degree = np.nanargmax(evidences)
regression = models[degree]
X_test = PolynomialFeature(degree=int(degree)).transform(x_test)
y, y_std = regression.predict(X_test, return_std=True)
plt.scatter(x_train, y_train, s=50, facecolor="none", edgecolor="steelblue", label="observation")
plt.plot(x_test, cubic(x_test), label="x(x-5)(x+5)")
plt.plot(x_test, y, label="prediction")
plt.fill_between(x_test, y - y_std, y + y_std, alpha=0.5, label="std", color="orange")
plt.legend()
plt.show()
plt.plot(evidences)
plt.title("Model evidence")
plt.xlabel("degree")
plt.ylabel("log evidence")
plt.show()